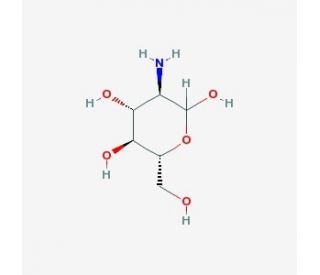
Struttura molecolare di D-Glucosamine, Numero CAS: 3416-24-8
D-Glucosamine (CAS 3416-24-8)
Numero CAS:
3416-24-8
Peso molecolare:
179.17
Formula molecolare:
C6H13NO5
Solo per uso in Ricerca. Non previsto per Uso Diagnostico o Terapeutico.
* Vedere Certificato di Analisi per informazioni sul lotto specifico (incluso il contenuto d'acqua).
LINK RAPIDI
Informazioni ordini
Descrizione
Informazione tecnica
Informazioni di sicurezza
SDS & Certificato d'analisi
La D-Glucosamina è un aminozucchero presente nel liquido delle articolazioni e nel tessuto connettivo. Svolge un ruolo fondamentale nella sintesi e nel mantenimento della cartilagine. È un elemento costitutivo fondamentale dei glicosaminoglicani (GAG) e dei proteoglicani, che sono componenti essenziali della struttura della cartilagine. Si ritiene che la D-Glucosamina stimoli la produzione di nuova cartilagine. La D-Glucosamina viene utilizzata come substrato in saggi enzimatici e studi metabolici per indagare varie vie biochimiche.
D-Glucosamine (CAS 3416-24-8) Referenze
- La D-glucosamina inibisce la proliferazione delle cellule tumorali umane attraverso l'inibizione di p70S6K. | Oh, HJ., et al. 2007. Biochem Biophys Res Commun. 360: 840-5. PMID: 17624310
- La D-glucosamina riduce l'HIF-1alfa attraverso l'inibizione della traduzione della proteina nelle cellule di cancro alla prostata DU145. | Park, JY., et al. 2009. Biochem Biophys Res Commun. 382: 96-101. PMID: 19254699
- La D-glucosamina promuove l'efficienza di trasfezione durante l'elettroporazione. | Igawa, K., et al. 2014. Biomed Res Int. 2014: 485867. PMID: 24678506
- La forma della D-glucosamina. | Peña, I., et al. 2014. Phys Chem Chem Phys. 16: 23244-50. PMID: 25255812
- Studi 1H NMR dell'interazione molecolare di D-glucosamina e N-acetil-D-glucosamina con la capsaicina in mezzi acquosi e non acquosi. | Higuera-Ciapara, I., et al. 2017. Carbohydr Res. 452: 6-16. PMID: 28992455
- Formulazione dell'anomerizzazione e della protonazione nella d-glucosamina, basata sull'1H NMR. | Virués, C., et al. 2020. Carbohydr Res. 490: 107952. PMID: 32114014
- Confronto tra l'acidità e l'affinità con gli ioni metallici della D-glucosamina e della N-acetil-D-glucosamina, uno studio DFT. | Kotena, ZM. and Fattahi, A. 2020. J Mol Graph Model. 98: 107612. PMID: 32302939
- Miglioramento della produzione di D-Glucosamina in Escherichia coli mediante il blocco di tre vie coinvolte nel consumo di GlcN e GlcNAc. | Li, P., et al. 2020. Mol Biotechnol. 62: 387-399. PMID: 32572810
- Studio della reazione acido-catalizzata tra 2-metil e 2-(2,2,2-tricloroetossi) gluco-[2,1-d]-2-ossazoline. Sintesi di un derivato pseudo-tetrasaccaridico macrociclico della d-glucosamina. | Pertel, SS., et al. 2021. Carbohydr Res. 499: 108230. PMID: 33429169
- Liquidi ionici aminoacidici catalizzano la d-glucosamina in derivati della pirazina: Spiegazioni dalla spettroscopia NMR. | Liu, P., et al. 2021. J Agric Food Chem. 69: 2403-2411. PMID: 33595305
- La d-glucosamina come ligando verde per la sintesi domino regio-stereoselettiva catalizzata da Cu(I) di (Z)-3-metileneisoindolina-1-oni e (E)-N-aril-4H-tiocromene-4-imine. | Singh, SK., et al. 2021. ACS Omega. 6: 21125-21138. PMID: 34423220
- Scissione anomerica stereoausiliaria del legame C-N della d-Glucosamina per la preparazione di imidazo[1,5-a]piridine. | Zeng, K., et al. 2022. Chemistry. 28: e202200648. PMID: 35319128
- Preparazione dell'acido solido poroso a base di carbonio dall'amido per un'efficiente degradazione del chitosano a D-glucosamina. | Wang, W., et al. 2022. Int J Biol Macromol. 209: 1629-1637. PMID: 35447270
- Produzione di d-glucosamina dall'idrolizzazione del chitosano su un catalizzatore acido solido derivato dal glucosio. | Zhang, H., et al. 2018. RSC Adv. 8: 5608-5613. PMID: 35542433
- Rettifica: Scissione anomerica stereoausiliaria del legame C-N della d-Glucosamina per la preparazione di imidazo[1,5-a]piridine. | Zeng, K., et al. 2022. Chemistry. 28: e202202181. PMID: 35852152
Inibitore di:
β-defensin 44, AGP-2, AMPKγ3, C20orf103, C20orf117, COL19A1, Eps15L1, Forssman Antigen, GFAT1, GFOD1, HAPLN1, HAS3, Hex, Hexokinase, L-ficolin, LYZL4, OSX, PGM 2L1, PGRP-Iβ, PIG-K, e Telethonin.Attivatore di:
α-mannosidase, β-1,3-Gal-T2, β-1,4-Gal-T5, β-1,4-Gal-T7, β-1,4-GalNAc-T3, β-4-Gal-T1, β-defensin 133, β3Gn-T2, β3Gn-T6, 4933434I20Rik, aggrecan 1, ALG1, ALG12, ALG13, ALG5, Annexin I, BC030867, C3orf21, C3orf64, C4ST-2, caspase-12, CD-MPR, CD209c, CD59, CEACAM3, CHAD, CHADL, chondroitin sulfate, chondroitin-4-sulfate, CHSY1, CILP, COL13A1, COL9A3, Collagen Type II, CSGlcA-T, D4ST, DPM2, DSCR 5, Endoglycan, EP58, EXT2, EXTL1, EXTL3, ficolin B, FLJ23356, FUCA1, G6PT, GBP11, GCNT4, GCS-α-1, GDF-16, GFAT1, GFAT2, GK1, GLCE, GlcNAc kinase, GLCNE, GLT25D2, glypican-2, GM2/GD2 Synthase, GMPPA, GNPDA1, GNPDA2, GnT-II, GnT-III, GnT-IVA, GnT-V, GPT, GXYLT2, HAPLN2, HS2ST1, IDS, LARGE, LYSMD2, MAN1A2, Matrilin-1, Matrilin-2, Matrilin-3, Matrilin-4, MBL-C, MPG1, Mucin 15, MXRA5, NAGA, NAGPA, NANS, NPL, Npl1, O-GlcNAc transferase, PGRP-S, PIG-N, PIG-preY, PKLR, PMM1, POMGnT1, RFT1, Sialyltransferase 7D, Sialyltransferase 7E, SLC35A3, SLC35F4, ST8Sia V, Stabilin-1, UAP1, UAP1L1, UDP-GlcDH, V-ATPase α1, V-ATPase D2, e XylT-II.Informazioni ordini
| Nome del prodotto | Codice del prodotto | UNITÀ | Prezzo | Quantità | Preferiti | |
D-Glucosamine, 1 g | sc-278917A | 1 g | $201.00 | |||
D-Glucosamine, 10 g | sc-278917 | 10 g | $779.00 |
