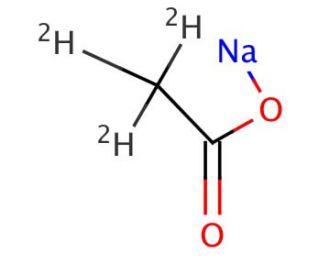
Sodium Acetate-D3 (CAS 39230-37-0)
LINKS RÁPIDOS
O Acetato de Sódio-D3 é frequentemente utilizado na investigação de espetroscopia de ressonância magnética nuclear (RMN) para investigar estruturas e dinâmicas moleculares. Este composto marcado isotopicamente permite uma maior sensibilidade de deteção em estudos de RMN, tornando-o útil para analisar moléculas orgânicas complexas. Os investigadores utilizam o Acetato de Sódio-D3 em estudos metabólicos para rastrear e quantificar as vias metabólicas em vários organismos. É também utilizado em estudos centrados nos mecanismos enzimáticos, particularmente os que envolvem reacções de transferência de acetilo. Além disso, o Acetato de Sódio-D3 é utilizado na investigação para estudar as interacções e a estabilidade das proteínas, fornecendo informações sobre a sua dobragem e função.
Sodium Acetate-D3 (CAS 39230-37-0) Referencias
- Duas doenças neurodegenerativas diferentes causadas por proteínas com estruturas semelhantes. | Mo, H., et al. 2001. Proc Natl Acad Sci U S A. 98: 2352-7. PMID: 11226243
- Concentração e separação de amostras para espetroscopia RMN de volume nanolitro utilizando isotacoforese capilar. | Kautz, RA., et al. 2001. J Am Chem Soc. 123: 3159-60. PMID: 11457037
- Caracterização dos citocromos c(552) de Hydrogenobacter thermophilus expressos no citoplasma e no periplasma de Escherichia coli. | Karan, EF., et al. 2002. J Biol Inorg Chem. 7: 260-72. PMID: 11935350
- Estrutura em solução e dinâmica da proteína de atividade inibitória do melanoma. | Lougheed, JC., et al. 2002. J Biomol NMR. 22: 211-23. PMID: 11991352
- O I-motif híbrido PNA-DNA: implicações para os contactos açúcar-açúcar na tetramerização do i-motif. | Modi, S., et al. 2006. Nucleic Acids Res. 34: 4354-63. PMID: 16936319
- A descarboxilação do uroporfirinogénio como referência para a competência catalítica das enzimas. | Lewis, CA. and Wolfenden, R. 2008. Proc Natl Acad Sci U S A. 105: 17328-33. PMID: 18988736
- Cristalização e experiência preliminar de difração de neutrões da farnesil pirofosfato sintase humana complexada com risedronato. | Yokoyama, T., et al. 2014. Acta Crystallogr F Struct Biol Commun. 70: 470-2. PMID: 24699741
- Avaliação da estrutura de ordem superior de Humira®, Remicade®, Avastin®, Rituxan®, Herceptin® e Enbrel® por impressão digital 2D-NMR. | Hodgson, DJ., et al. 2019. J Pharm Biomed Anal. 163: 144-152. PMID: 30296716
- Avaliação e otimização de métodos de manuseamento de amostras para quantificação de ácidos gordos de cadeia curta em amostras de fezes humanas por GC-MS. | Hsu, YL., et al. 2019. J Proteome Res. 18: 1948-1957. PMID: 30895795
- Conformações de compostos modelo de proteínas. I. N-metilamida de acetilglicina. | Koyama, Y. and Shimanouchi, T. 1968. Biopolymers. 6: 1037-76. PMID: 5663406
- A cardiolipina modula a estrutura secundária do péptido de pré-sequência da subunidade IV da citocromo oxidase: um estudo 2D 1H-NMR. | Chupin, V., et al. 1995. FEBS Lett. 373: 239-44. PMID: 7589474
- Estrutura em solução do domínio de ligação a Ras do c-Raf-1 e identificação da sua superfície de interação com Ras. | Emerson, SD., et al. 1995. Biochemistry. 34: 6911-8. PMID: 7766599
- Ligação de álcool a lipossomas por RMN de 2H e ensaios de ligação com marcadores de rádio: a partição descreve a ligação? | Dubey, AK., et al. 1996. Biophys J. 70: 2307-15. PMID: 9172754
- Mimetização estrutural de uma proteína nativa por um domínio de ligação minimizado. | Starovasnik, MA., et al. 1997. Proc Natl Acad Sci U S A. 94: 10080-5. PMID: 9294166
Informacoes sobre ordens
| Nome do Produto | Numero de Catalogo | UNID | Preco | Qde | FAVORITOS | |
Sodium Acetate-D3, 5 g | sc-264316 | 5 g | $99.00 |
